A Snippet for Error Handling in R: An Example with ASReml-R
Dr. Giovanni Galli
12 January 2024
Automating analytical workflows is a common procedure that allow us to let the computer do the hard work. When that is the case, we must prepare our system to be robust enough so it can anticipate and handle errors that would break the pipeline, causing delays or even reporting undetected incorrect outcomes. This is particularly important when dealing with large or many datasets, complex and unbalanced statistical procedures, or when there are too many repeated tasks to be performed (e.g., many statistical models to be tested).
Let’s dig a bit on this topic within R by having a look at a simple example showing how to handle errors and reporting why things fail. For this example we are going to use ASReml-R, but this principle is transferable to other code, packages, and even programming languages.
We will start by loading the ASReml-R library:
Let’s create some ASReml-R calls in a character object that we will evaluate later.
The first call is:
And the second call is:
Next, we will fit something that is a bit complex (spatial analysis with nugget) given the data at hand:
Now let’s create our own function (called error_handling_asreml) that will: execute an ASReml call, check if the models have converged (update if not and stop after some tries), and finally return the fitted models or an error message. Note that error handling in R does not require a wrapping function, but it is a good way to keep things organized. The purpose of each part of the function is explained in the comments.
#' @title Error handling ASReml-R
#'
#' @param call is the asreml call to be evaluated provided as character.
#' @param max_tries is the maximum number of tries to update the model if it
#' has not converged yet.
#'
#' @return A list with the exit status and the model object if it converged.
error_handling_asreml <- function(call = NULL, max_tries = 5) {
# Inform that the function has started.
message("MESSAGE: initializing asreml...\n")
# Create new environment to hold error messages. This can also go outside the
# function to make it more general.
ErrorEnv <- new.env()
# The tryCatch function is the core of error handling. If all is good with
# the evaluated expression, it will move on, if not,
# it will collect the error message into the error environment.
tryCatch(
# This is where we run our code that might fail. Here we are evaluating
# the call to ASReml-R. The eval(parse(text)) is used to evaluate
# the character object as a function call.
expr = eval(parse(text = paste0("mod<-", call))),
# And this is where we handle things when they go wrong. Notice we are
# saving (assigning) the error message into the error environment.
error = function(e){
assign("error_message", conditionMessage(e), envir = ErrorEnv)
})
# If the model did not run (does not exist because the evaluation failed),
# we can return the error message and stop here.
if (!exists("mod")) {
# This message will be printed in the console so we know it failed.
message("ERROR: see $exit for more information.")
# And this will be saved in the exit object for later debugging.
return(list(
exit = paste0("ASReml-R unable to run the call: \n\n", call, "\n\n",
"Collected error message: ", ErrorEnv$error_message))
)
}
# Assuming the model runs, you can check if it converged and give it a few
# more tries in case it has not. This part is optional, but it is a good way
# to ensure parameter and likelihood convergency.
# Let's initialize a counter.
cur_try = 1
# Using the while loop to keep trying until it converges or max_tries is met.
while (!mod$converge) {
# Report update.
message("MESSAGE: base model likelihood has not converged. updating.\n")
# Call tryCatch.
tryCatch(
# Execute "update" function. If there is no required change to the call
# this code can be used for any model.
expr = eval(parse(text = 'mod<-update(mod)')),
# And again, this is where we handle things when they go wrong.
error = function(e){
assign("error_message", conditionMessage(e), envir = ErrorEnv)
})
# Report and save error message in case it failed.
if (is.null(mod)) {
message("ERROR: see $exit for more information.")
return(list(
exit = paste0("ASReml-R unable to update the call: \n\n", call, "\n\n",
"Collected error message: ", ErrorEnv$error_message),
)
)
}
# If the model did not converge after a number of tries,
# we should break and inform.
if (cur_try == max_tries) {
return(list(exit = "Model did not converge after several updates!"))
# Update try counter for another iteration.
cur_try = cur_try + 1
} # End tries.
} # End while.
# If all is OK, here is where you'll add you call to wald()
# predict() or other complementary code. You can wrap it in a tryCatch() as
# we did before to make things more manageable and stable.
# Return results.
message("MESSAGE: model converged.\n")
return(list(exit = "converged", model = mod))
}Let’s execute the error_handling_asreml function in batch mode with lapply and see what happens.
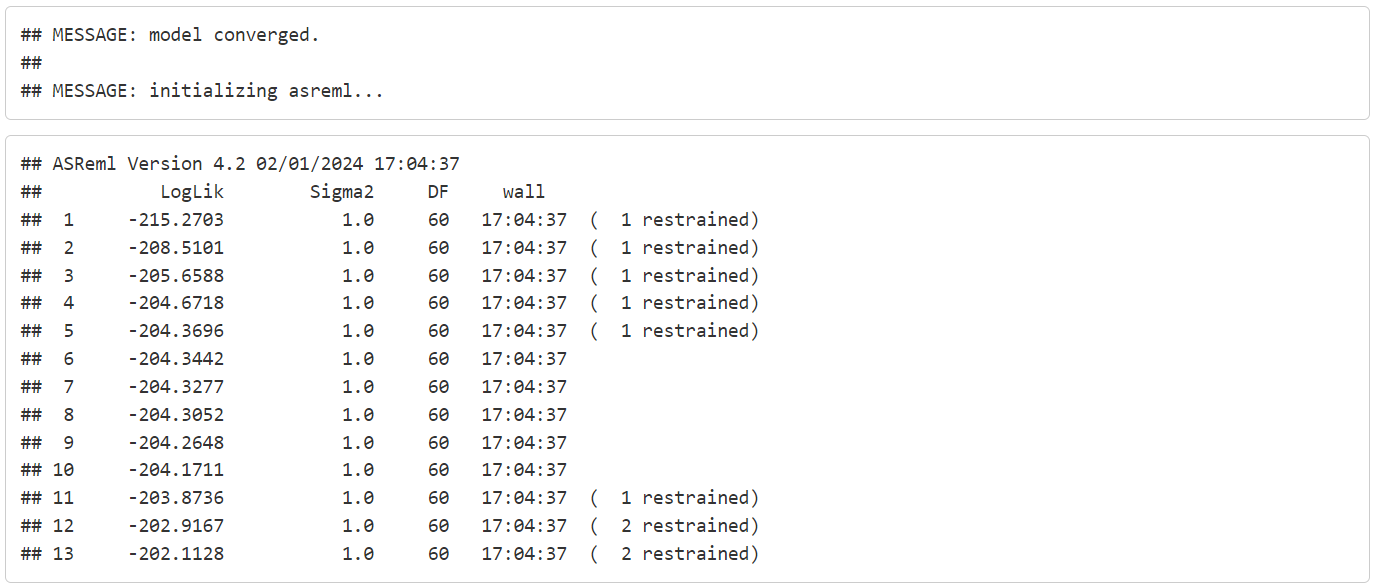

We can see that call1 failed, call2 succeeded, and call3 succeeded but needed additional updates to converge. Since we collected the error message we can have a look at what went wrong with call1: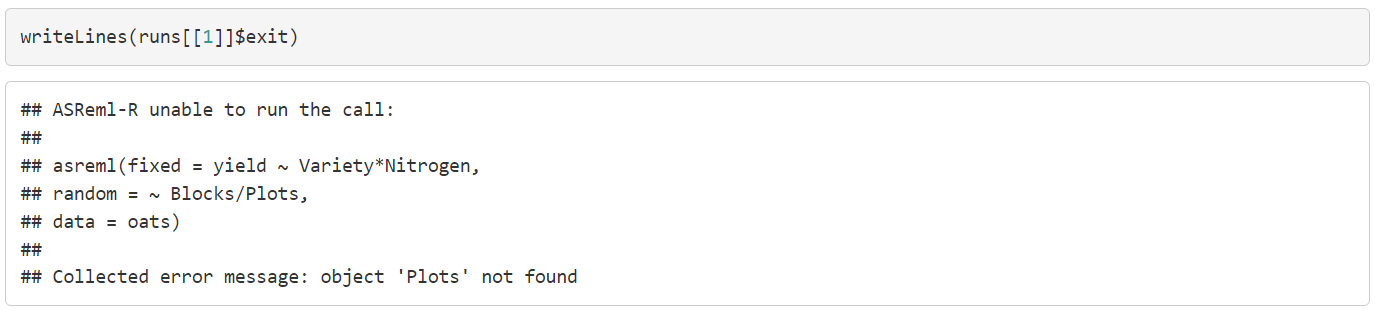
Note that the model did not run because the factor Plots does not exist as the actual name is Wplots. Having the error message stored can assist us in fixing the issue and this does not get lost if other messages are printed in the console.
We hope this gives you an idea and basic template on how to handle errors in R. There are many other ways of doing this, and you must adapt the code to your needs. Download here the full R file with the code.
About the author
Dr. Giovanni Galli is an Agronomist with an M.Sc. and Ph.D. in Genetics and Plant Breeding from the University of São Paulo (USP)/Luiz de Queiroz College of Agriculture (ESALQ). He currently works as a Statistical Consultant at VSN International, United Kingdom. Dr. Galli has experience in field trials, quantitative genetics, conventional and molecular breeding (genomic prediction and GWAS), machine learning, and high-throughput phenotyping.
Popular
Related Reads
